About PyMeta.com
PyMeta is an online Meta-analysis tool website. It was created and supported with Python, a strong and amazing computer language.
This web-based application was designed to perform some Evidence-based medicine (EBM) tasks, such as:
- Combining effect measures (OR, RR, RD for count data and MD, SMD for continuous data);
- Heterogeneity test(Q/Chi-square test);
- Subgroup analysis;
- Cumulative meta-analysis;
- Sensitivity analysis (one or two factors);
- Plots drawing: forest plot, funnel plot, and bar-line, cross-block plot.
Statistical algorithms in this software cited from:
Jonathan J Deeks and Julian PT Higgins, on behalf of the Statistical Methods Group of The Cochrane Collaboration. Statistical algorithms in Review Manager 5, August 2010.
Our web pages were well-compatible to handhold sets, e.g. android phones and iPhone, which means you can access this tool via mobile phone anywhere anytime, enjoy!
This is an ongoing project, so, any questions and suggestions from you are very welcome.
Please contact me with email, or say some words in
Guestbook.
Please cite me in any publictions like:
Deng Hongyong. PythonMeta, Python module of Meta-analysis, cited 2018-07-09 (or your time); 1 screen(s). Available from URL: http://www.pymeta.com
Thank you.
Go Top
How to input data?
You can type or paste your studies into the data-input textarea.
Each study in one line, like:
study name, e1, n1, e2, n2
for binary data:
e1,n1: event counts (e.g. ) and sample size of experiment group;
e2,n2: event counts and sample size of control group.
e.g.
Fang 2015, 15, 40, 24, 37
Gong 2012, 10, 40, 18, 35
Liu 2015, 30, 50, 40, 50
Long 2012, 19, 40, 26, 40
Pan 2015a, 57, 100, 68, 100
Wang 2001, 13, 18, 17, 18
Wang 2003, 7, 86, 15, 86
#This is a sample of binary data.
#Input one study in a line;
#Syntax: study name, e1, n1, e2, n2
#e1,n1: events and sample size of experiment group;
#e2,n2: events and sample size of control group.
or
study name, m1, sd1, n1, m2, sd2, n2
for continuous data:
m1, sd1, n1: mean, SD and sample size of experiment group;
m2, sd2, n2: mean, SD and sample size of control group.
e.g.
Atmaca 2005, 20.9, 6.0, 15, 27.4, 8.5, 14
Guo 2014, 12.8, 5.2, 51, 11.9, 5.3, 51
Liu 2010, 23.38, 5.86, 35, 24.32, 5.43, 35
Wang 2012, 15.67, 8.78, 43, 18.67, 9.87, 43
Xu 2002, 15.49, 7.16, 50, 21.72, 8.07, 50
Zhao 2012, 12.8, 5.7, 40, 13.0, 5.2, 40
#This is a sample of continuous data.
#Input one study in a line;
#Syntax: study name, m1, sd1, n1, m2, sd2, n2
#m1, sd1, n1: mean, SD and sample size of experiment group;
#m2, sd2, n2: mean, SD and sample size of control group.
Tags like '
<subgroup>name=subgroup_name' can mark all studies above as a subgroup.
And a tag of '
<nototal>' can hide overall result of subgroup analysis.
e.g.
Fang 2015,15,40,24,37
Gong 2012,10,40,18,35
Liu 2015,30,50,40,50
Long 2012,19,40,26,40
Wang 2003,7,86,15,86
<subgroup>name=short term
Chen 2008,20,60,28,60
Guo 2014,31,51,41,51
Li 2015,29,61,31,60
Yang 2006,21,40,31,40
Zhao 2012,27,40,30,40
<subgroup>name=medium term
<nototal>
#This is a sample of subgroup.
#Cumulative meta-analysis and Senstivity analysis will ignore all <subgroup> tags.
#And you can add a line of <nototal> to hide the Overall result.
Go Top
Output plots
Forest plot
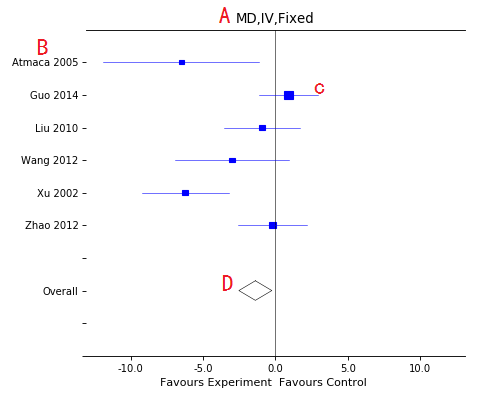
Forest plot
A, Title of the plot with some information:
- Effect measure: MD-Mean difference,SMD-Standard mean difference,RR-Risk ratio, OR-Odds ratio, RD-Ratio difference;
- Algorithm: IV-Inverse variance,MH-Mantel Haenszel,Peto;
- Effect models: Fixed or random models;
B, Included studies list;
C, Each study's effect, include CI line and central block(position for effect and size for weight);
D, Overall effect diamond, empty for high heterogeneity (I-square more than 50%) and filled for lower heterogeneity.
Forest plot with subgroup
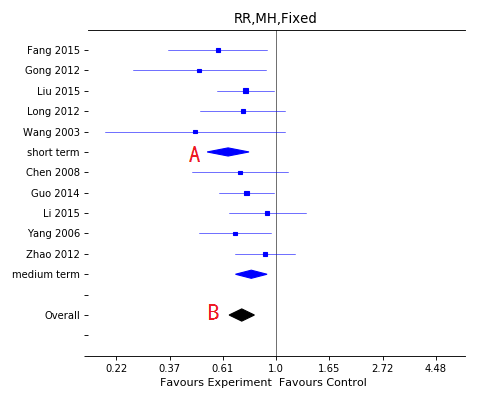
Forest plot with subgroup
A, Subgroup effect;
B, Overall effect.
Funnel plot

Funnel plot
A, Scatter dots of studies;
B, Boundary lines of effect;
C, Overall effect line.
An Egger's test will be performed only if included studies more than (or equal to) 10, and the results show on the top right.
Forest plot of cumulative meta-analysis
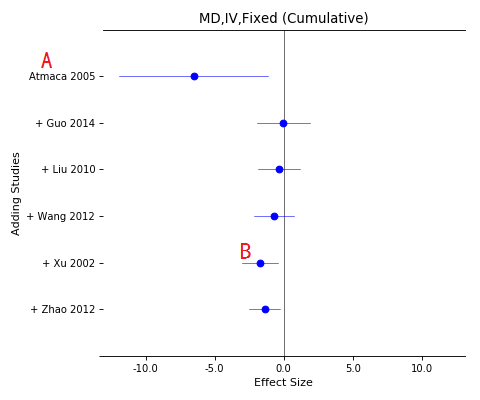
Forest plot of cumulative meta-analysis
A, The cumulative studies list (downward);
B, Total effects while each study added in the pool.
Polar_forest plot of sensitivity meta-analysis
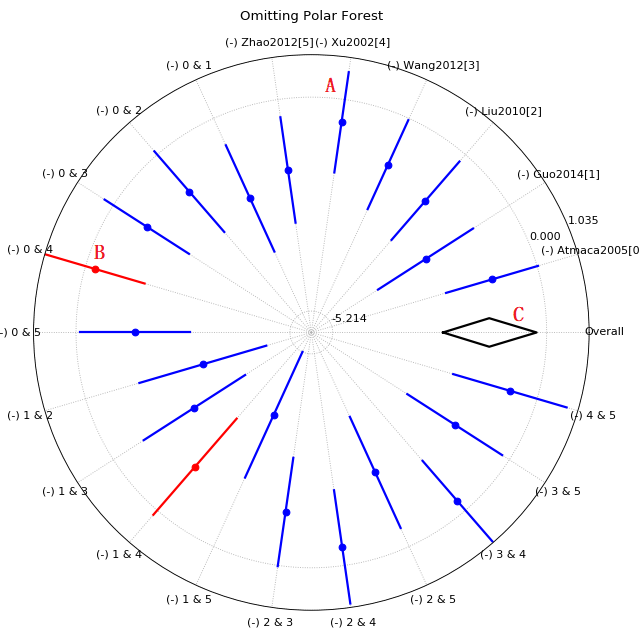
Polar_forest plot of sensitivity meta-analysis
This kind of figure is designed for reviews with large amounts of trials, and map the normal forest plot into a polar plot.
A, Effect while one or two trial(s) be removed, blue color means the I-square are still higher than 50%;
B, Red line for those I-square decreased to below 50% while one(or two) study removed;
C, Overall effect diamond (without any trial removed), again, empty for high heterogeneity and filled for lower heterogeneity.
Bar_line of sensitivity meta-analysis
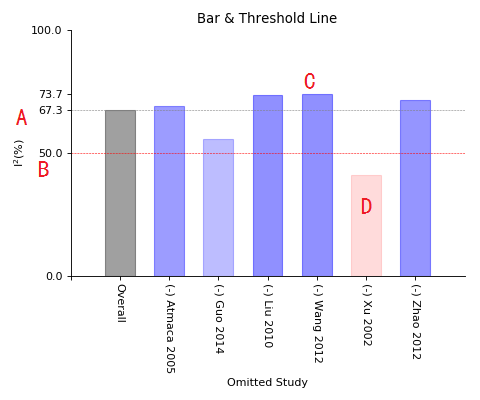
Bar_line of sensitivity meta-analysis
A, I-square value of overall test;
B, 50% I-square line ('50%' always regarded as threshold value, lower means fewer heterogeneity and higher means high heterogeneity);
C, I-square bar, grey for overall, blue for those one(or two) study removed, but I-square still higher than 50%;
D, Red bar for those I-square decreased to below 50% while one(or two) study removed.
Colored cross block of sensitivity meta-analysis
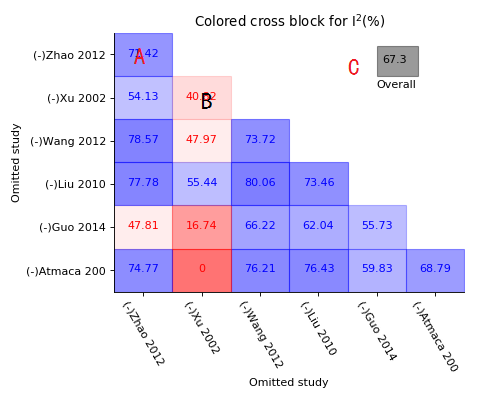
Colored cross block of sensitivity meta-analysis
A, Each block shows the I-square value of which two crossed studies were removed, blue block for those I-square still higher than 50% after removing;
B, Red block for those I-square decreased to below 50% while two studies removed;
C, Grey for overall I-square.
Go Top
DISCLAIMER
No warranties
PyMeta.com is provided "as is" without any representations or warranties, expressed or implied. PyMeta.com makes no representations or warranties in relation to this website or the information and materials provided on this website.
Without prejudice to the generality of the foregoing paragraph, PyMeta.com does not warrant that: this website will be constantly available, or available at all; or the information on this website is complete, true, accurate or non-misleading.
Nothing on this website constitutes, or is meant to constitute, advice of any kind. (If you require advice in relation to any (legal, financial or medical) matter you should consult an appropriate professional.)
Limitations of liability
PyMeta.com will not be liable to you (whether under the law of contract, the law of torts or otherwise) in relation to the contents of, or use of, or otherwise in connection with, this website:
for any indirect, special or consequential loss; or
for any business losses, loss of revenue, income, profits or anticipated savings, loss of contracts or business relationships, loss of reputation or goodwill, or loss or corruption of information or data.
These limitations of liability apply even if PyMeta.com has been expressly advised of the potential loss.
Reasonableness
By using this website, you agree that the exclusions and limitations of liability set out in this website disclaimer are reasonable.
If you do not think they are reasonable, you must not use this website.
Other parties
(You accept that, as a limited liability entity, PyMeta.com has an interest in limiting the personal liability of its officers and employees. You agree that you will not bring any claim personally against PyMeta.com's officers or employees in respect of any losses you suffer in connection with the website.)
(Without prejudice to the foregoing paragraph,) you agree that the limitations of warranties and liability set out in this website disclaimer will protect PyMeta.com's officers, employees, agents, subsidiaries, successors, assigns and sub-contractors as well as PyMeta.com.
Unenforceable provisions
If any provision of this website disclaimer is, or is found to be, unenforceable under applicable law, that will not affect the enforceability of the other provisions of this website disclaimer.
Go Top






